Research
Rationale
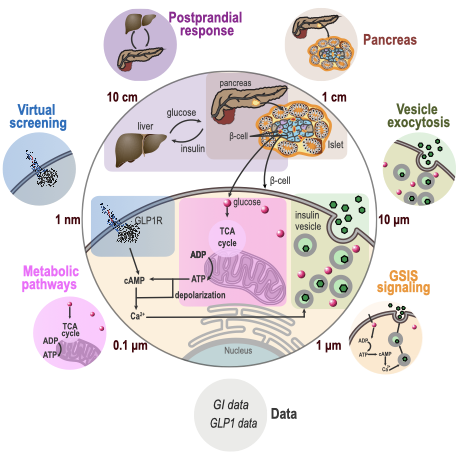
[Show legend] Illustration of varied experimental datasets and models of different aspects (small outer circles) of the human pancreatic β-cell (large inner circle). Insulin production and secretion is coordinated by a multiscale network of signals from the body, pancreas, and within the β-cell. Each of these aspects has been modeled separately, including coarse-grained spatiotemporal Brownian Dynamics simulation of insulin vesicle exocytosis; a molecular network model of glucose-stimulated insulin signaling (GSIS); a network model of metabolic pathways; an atomic structural model of glucagon-like peptide-1 receptor (GLP1R) activation via virtual screening; a linear model of the pancreatic cell population; Ordinary Differential Equations for systemic insulin postprandial response; and data on glucose intake (GI) rate upon food ingestion and GLP1 plasma concentration.
The cell is difficult to model because it consists of many different components that self-organize spatiotemporally. Computational modeling in general can be understood as a search for models that are sufficiently consistent with input information about the modeled system1. Given the complexity of the cell, computing an accurate, precise, and complete cell model requires a vast amount of information. This information can in principle be provided by different types of experimental datasets (eg, a cryo-electron tomography map of the cell and a large-scale affinity co-purification dataset), physical principles (eg, statistical mechanics of protein structure and dynamics), statistical preferences (eg, a statistical potential extracted from known protein structures), and prior models of the cell and/or its parts (eg, protein structure models and molecular signaling networks)2. Moreover, these experimental datasets and models must by necessity be produced by many scientists with diverse expertise, working iteratively with one another over long periods of time. Such long-term, collaborative, and interdisciplinary work requires experimental datasets and models to be archived, shared among the collaborators, analyzed, used for further modeling, and disseminated to the broader community. In other words, cell modeling is data-centric: advances are driven by the collaborative creation, validation, curation, and exchange of complex data3. As a result, a cyberinfrastructure for comprehensive data management4 is critical for a successful cell mapping project. This cyberinfrastructure needs to be efficient, robust, and extensible, including by linking with existing external resources into a federated system5, wherever possible. In addition, it must be freely available, so as to encourage its adoption, further development, and use by as many scientists as possible, in turn making it more useful to all.
Goal
Our goal is to develop CellLab, a comprehensive data-centric cyberinfrastructure that facilitates mapping the architecture and function of the cell. Most importantly, CellLab supports a large team of collaborators to store, analyze, use, share, and disseminate many varied data, including experimental datasets and cell models, in multiple iterations of experiment and modeling that are required for cell mapping. CellLab promotes collaboration and reproducibility by integrating data management with various modeling engines and modeling workflows. Thus, CellLab creates a data-centric, socio-technical ecosystem for collaborative science by providing a framework for sharing expertise, experimental data, models, and other resources.
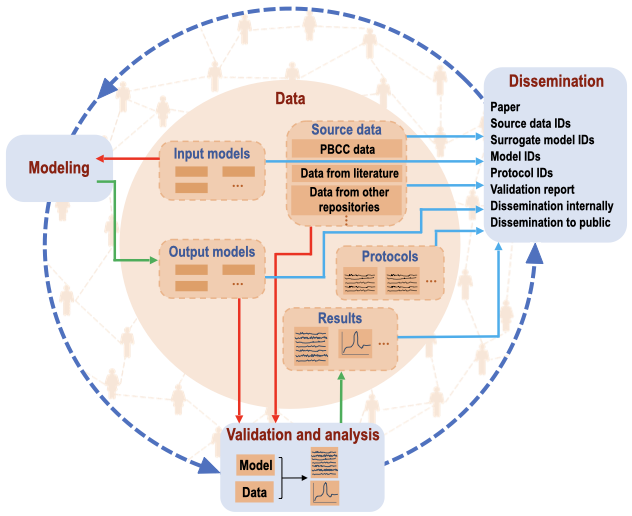
[Show legend] Data-centric approach to cell modeling. The central circle shows the data under management (dark orange rectangles). The data include the experimental datasets from the PBCC, literature, and other existing repositories, input models from external resources, output models, modeling and analysis parameters and protocols, and results produced in CellLab. The outer circle shows the processes of modeling, validation, and dissemination. The dashed orange outline of each box inside the data circle represents the data flow between the different categories. Red arrows represent data export for modeling, validation, and analysis. Green arrows show the input of processed or modified data. Cyan arrows represent data links via unique identifiers for dissemination. Blue dashed arrows between three major modules represent the scientific process iterating through experiment and modeling. People icons in the background indicate the individuals, labs, and communities interacting with CellLab.
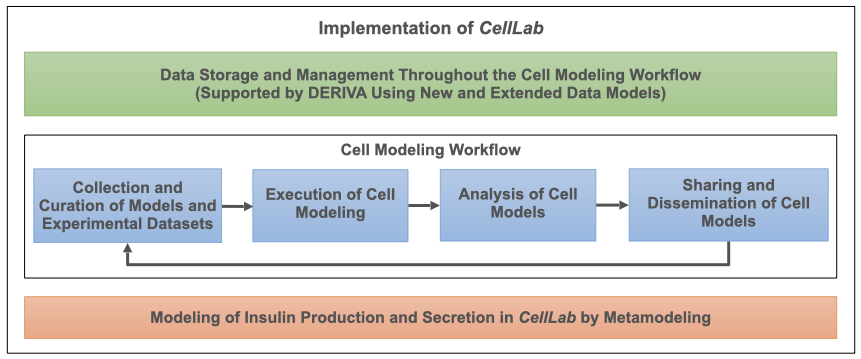
[Show legend] CellLab workflow. The green box corresponds to the CellLab infrastructure that supports the iterative and evolving cell modeling workflow (blue boxes). The red box indicates the first modeling application (use case) implemented in CellLab. Modeling applications relying on other modeling methods will be implemented in the future.
Data-centric approach to cell mapping
CellLab is built using our scientific asset management platform Deriva6. Data management abides by the FAIR (Findable, Accessible, Interoperable, and Reusable) principles7. To drive the development of CellLab, we use it for modeling glucose-stimulated insulin production and secretion8, one of the key functions of the β-cell, in collaboration with the Pancreatic β-Cell Consortium (PBCC). This use case challenges all aspects of CellLab, because insulin production and secretion encompass many of the complexities of the whole cell, including aspects best described using different types of experimental data and models at different scales. Thus, our existing open-source automated Bayesian metamodeling9 is the first instantiation of a modeling method in CellLab; metamodeling updates a set of heterogeneous input models, such as spatiotemporal and network models, to make them more consistent with each other. Iterative application of CellLab to the use case results in rapid improvement of CellLab's user-centered design. Additional use cases are under development, including one from the Cancer Cell Map Initiative (CCMI).

[Show legend] System architecture of CellLab. The main components consist of a Deriva catalog that is customized by an evolving CellLab information model. The catalog is used to organize and describe all CellLab computational experiments and associated assets, including modeling results (stored in a cloud-hosted object store) and cell modeling programs. Data is extracted and inserted into CellLab via citable, reproducible data collection or open programmatic interfaces. Data are organized for cell modeling experiments by creating an evolving collection of CellLab data models, which enable data discovery and sharing. Curated data can be transferred in and out of CellLab as citable datasets for purposes of executing modeling and analysis as well as import and export to external repositories. Model execution may take place on a local workstation, a cloud-hosted platform such as WholeTale, and on-demand cloud-hosted services.
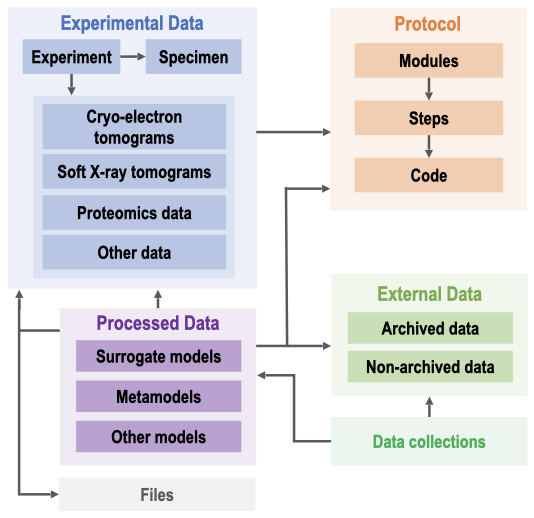
[Show legend] A high-level representation of the data model for CellLab. The data model supports the storage and management of different types of experimental data as well as processed data, including input and output models. Metadata information regarding data from external resources, file formats, data processing protocols, and associated software code are also described. The boxes are grouped and color-coded according to the information captured and the arrows correspond to the relationships (eg, foreign keys) between different elements in the model. A more detailed data schema is also available.
- Sali, A. From integrative structural biology to cell biology. Journal of Biological Chemistry 100743 (2021). doi:10.1016/j.jbc.2021.100743
- Singla, J., McClary, K. M., White, K. L., Alber, F., Sali, A. & Stevens, R. C. Opportunities and Challenges in Building a Spatiotemporal Multi-scale Model of the Human Pancreatic β Cell. Cell 173, 11–19 (2018).
- Leonelli, S. Data-Centric Biology. (University of Chicago Press, 2016).
- Roth, Y. D., Lian, Z., Pochiraju, S., Shaikh, B. & Karr, J. R. Datanator: an integrated database of molecular data for quantitatively modeling cellular behavior. Nucleic Acids Res. 49, D516–D522 (2021).
- Vallat, B., Webb, B., Westbrook, J. D., Sali, A. & Berman, H. M. Development of a Prototype System for Archiving Integrative/Hybrid Structure Models of Biological Macromolecules. Structure 26, 894–904 e2 (2018).
- Schuler, R. E., Kesselman, C. & Czajkowski, K. Accelerating data-driven discovery with scientific asset management. in 2016 IEEE 12th International Conference on e-Science (e-Science) (2017). doi:10.1109/eScience.2016.7870883
- Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J. W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., Gonzalez-Beltran, A., Gray, A. J. G., Groth, P., Goble, C., Grethe, J. S., Heringa, J., t Hoen, P. A. C., Hooft, R., Kuhn, T., Kok, R., Kok, J., Lusher, S. J., Martone, M. E., Mons, A., Packer, A. L., Persson, B., Rocca-Serra, P., Roos, M., van Schaik, R., Sansone, S. A., Schultes, E., Sengstag, T., Slater, T., Strawn, G., Swertz, M. A., Thompson, M., Van Der Lei, J., Van Mulligen, E., Velterop, J., Waagmeester, A., Wittenburg, P., Wolstencroft, K., Zhao, J. & Mons, B. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, (2016).
- Fu, Z., Gilbert, E. R. & Liu, D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr. Diabetes Rev. 9, 25–53 (2013).
- Raveh, B., Sun, L., White, K. L., Sanyal, T., Tempkin, J., Zheng, D., Bharath, K., Singla, J., Wang, C., Zhao, J., Li, A., Graham, N. A., Kesselman, C., Stevens, R. C. & Sali, A. Bayesian metamodeling of complex biological systems across varying representations. Proc. Natl. Acad. Sci. U. S. A. 118, (2021).